I-PV: Interactive Protein Sequence Visualization
Latest Version: 1.45
Examples of this version:
NF-κB-1 TNF-α P53 BRCA1
Media:
GIFS Videos Youtube
I have been extremely busy with the MutaFrame project which aims to be a platform to offer data visualization for variety of predictors.
I have written an entire visualization library from ground up called 'Lexicon' to handle visualization of data points well over 10000 points.
I would be gratefull if you check these projects and give me feedback on their feasibility. Meanwhile I am releasing a new version of I-PV with an entire new interface built to deal with mutations.
This new add-on is called INDORIL and it visualizes mutations on 3D cartesian coordinates. I have built an 3d svg renderer about 2 years ago, and never had the chance to trim it down and make it better.
I recently had bit of opportunity to revisit this unfinished work and make it slighltly better. The renderer can handle well above 1000 svg polygons at 60fps.
As again, I would be grateful if you let me know your first impressions about the tool. You can also check some new tutorials that I uploaded at youtube.
NF-κB-1
TNF-α
P53
BRCA1
DOWNLOAD v1.45
With the new 1.44 version being tested. I wanted to add two examples of this version. The new version encapsulates EGFR and JAK2, which I updated with the most recent data from Biomart.
I have decided to change the format of this page so that the more you scroll down you will also be able to see previous versions and news related to them. The v1.44 is a major overhaul to many features plus several bug fixes. For instance the placement of the deletions are changed for tiles. The main treat of I-PV v1.44 is the ability to change and modify colors. You can do this on desktop/iOS/android. Below you will find an introductory video for v1.44 and one example. Although formal download link is not generated, you can download the v1.44 from here or here. Be sure the check the source code at Github as well. I will try to update the changelog whenever possible.
Changing Colors with I-PV EGFR JAK2 DOWNLOAD v1.44 This is the official page for i-PV. Here, you will be able to download the software and also find a link to the blog site where additional materials are posted (or other softwares that I am currently working on). The new version of i-PV is ready and downloadable. I have made an introductory video below showing how to generate these graphs and also how to use the new lotus view. I had to trim the quality of the video for size constraints: Lotus view and the Introductory Video You can also check the GIFs page to check through a list of gifs highlighting features of I-PV. The version v1.43 is the first release of i-PV after its publication at Bioinformatics. I have added 4 examples outputs of version 1.43.2:
FOXP2 MYOSIN-2 TUBA1A TUBB2B
Let's do a small exercise to get you familiar with some of the features. First download a local version of the FOXP2 and this file. Try to plot the file and hover on the highlight elements to see their values. If you try to plot on the online version, you might have to hover on the region where the highlight is and click on the minimize (minus) icon to bring them back. This a recent bug introduced by chrome, however the offline version and the version 1.43.2 works ok. Howering on different highlight elements will reveal their information which should give you an idea about the data format. Follow the GIF below to see how the plotting is done. Now, hover on one of the highlight elements and click on the 'x' mark to remove them. Next try downloading this and plotting it on the same FOXP2 graph. You have just plotted the mRNA sequence of FOXP2.
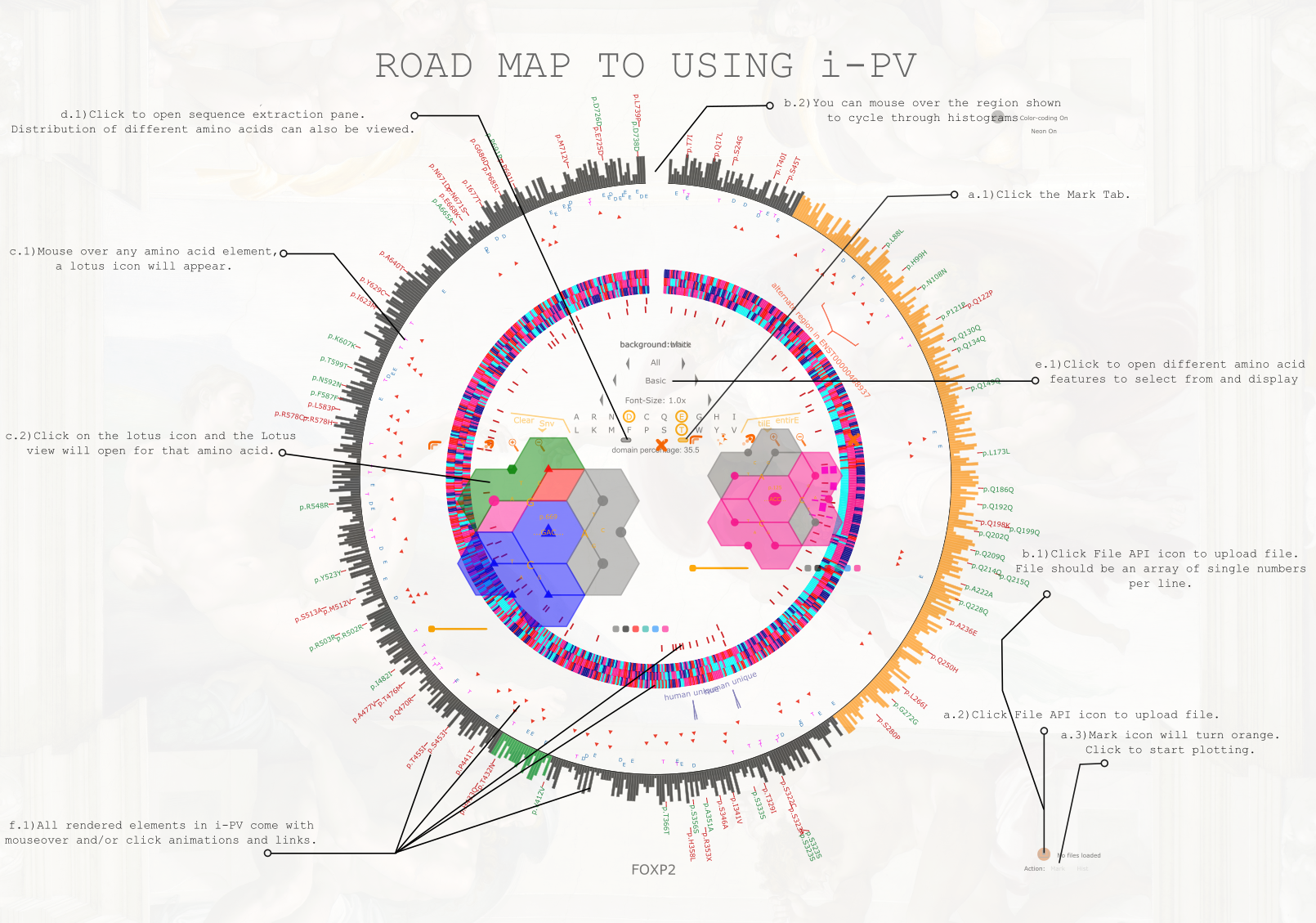
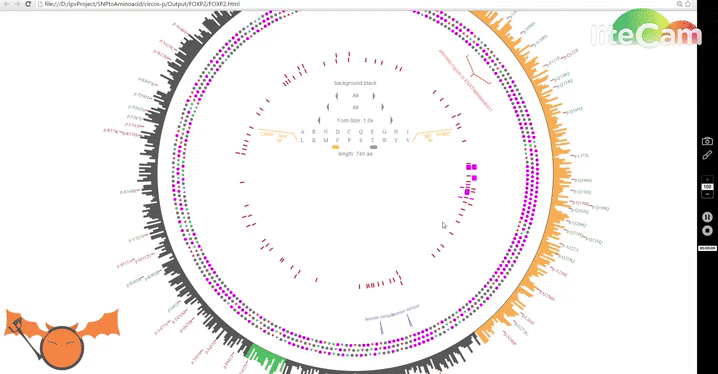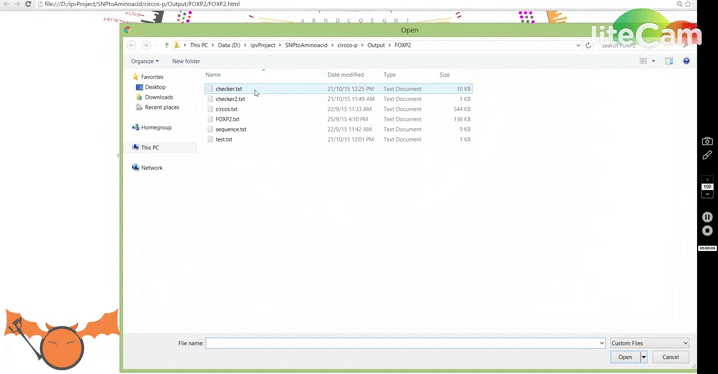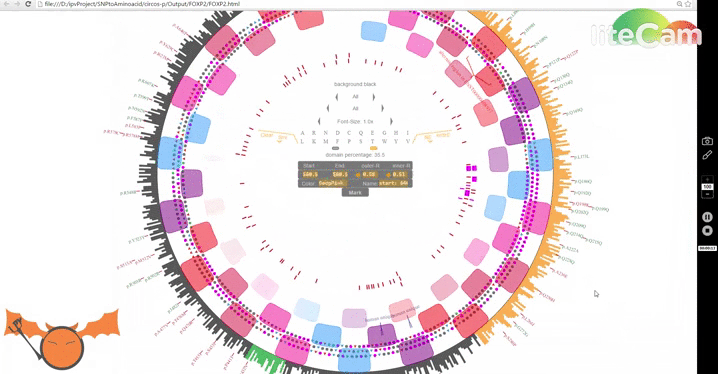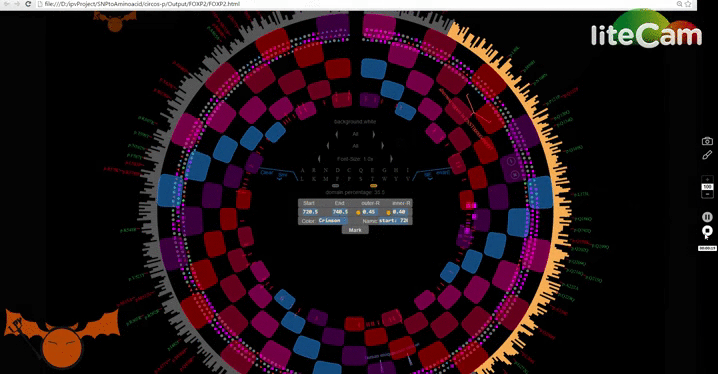 With this major update, I have added new features that incorporate mRNA data as well, and a new mode to visualize amino acid changes: Lotus-View. For now I have prepared a road map
to demonstrate the usage of i-PV, including the new visual mode. Below is the low resolution road-map. Send me an email if you need the high resolution one.
With this major update, I have added new features that incorporate mRNA data as well, and a new mode to visualize amino acid changes: Lotus-View. For now I have prepared a road map
to demonstrate the usage of i-PV, including the new visual mode. Below is the low resolution road-map. Send me an email if you need the high resolution one.
 I-PV is a software that uses Circos to render interactive images for protein sequence visualization. The goal of i-PV is to summarize
most sequence related data in one interactive image that is easy to generate and share. To have an idea of what i-PV can accomplish,
check out the sample outputs below. All elements you see on a typical i-PV output are interactable. For instance, the BRCA2 figure includes over 21000 animatable elements! These outputs are from the version 1.42-beta-5, meaning that they do not have lotus-view.
I-PV is a software that uses Circos to render interactive images for protein sequence visualization. The goal of i-PV is to summarize
most sequence related data in one interactive image that is easy to generate and share. To have an idea of what i-PV can accomplish,
check out the sample outputs below. All elements you see on a typical i-PV output are interactable. For instance, the BRCA2 figure includes over 21000 animatable elements! These outputs are from the version 1.42-beta-5, meaning that they do not have lotus-view.
BRCA1 CALR CFTR CDH1 BRCA2
To summarize the overall features of i-PV, a video is available on the blog site. Looking at the overall features is a good idea to have an overall understanding of how i-PV works:
Video-1 Video-2 Video-3 Video-4
To download i-PV, click the buttons below:
DOWNLOAD v1.43.1
or the recent bug fix:
DOWNLOAD v1.43.2
I have decided to change the format of this page so that the more you scroll down you will also be able to see previous versions and news related to them. The v1.44 is a major overhaul to many features plus several bug fixes. For instance the placement of the deletions are changed for tiles. The main treat of I-PV v1.44 is the ability to change and modify colors. You can do this on desktop/iOS/android. Below you will find an introductory video for v1.44 and one example. Although formal download link is not generated, you can download the v1.44 from here or here. Be sure the check the source code at Github as well. I will try to update the changelog whenever possible.
Changing Colors with I-PV EGFR JAK2 DOWNLOAD v1.44 This is the official page for i-PV. Here, you will be able to download the software and also find a link to the blog site where additional materials are posted (or other softwares that I am currently working on). The new version of i-PV is ready and downloadable. I have made an introductory video below showing how to generate these graphs and also how to use the new lotus view. I had to trim the quality of the video for size constraints: Lotus view and the Introductory Video You can also check the GIFs page to check through a list of gifs highlighting features of I-PV. The version v1.43 is the first release of i-PV after its publication at Bioinformatics. I have added 4 examples outputs of version 1.43.2:
FOXP2 MYOSIN-2 TUBA1A TUBB2B
Let's do a small exercise to get you familiar with some of the features. First download a local version of the FOXP2 and this file. Try to plot the file and hover on the highlight elements to see their values. If you try to plot on the online version, you might have to hover on the region where the highlight is and click on the minimize (minus) icon to bring them back. This a recent bug introduced by chrome, however the offline version and the version 1.43.2 works ok. Howering on different highlight elements will reveal their information which should give you an idea about the data format. Follow the GIF below to see how the plotting is done. Now, hover on one of the highlight elements and click on the 'x' mark to remove them. Next try downloading this and plotting it on the same FOXP2 graph. You have just plotted the mRNA sequence of FOXP2.


BRCA1 CALR CFTR CDH1 BRCA2
To summarize the overall features of i-PV, a video is available on the blog site. Looking at the overall features is a good idea to have an overall understanding of how i-PV works:
Video-1 Video-2 Video-3 Video-4
To download i-PV, click the buttons below:
DOWNLOAD v1.43.1
or the recent bug fix:
DOWNLOAD v1.43.2
LISENCE:
I retain the copyrights of Javascript used within this website and I-PV. So if you use parts of the code please give proper credit in a visible section of your project. The output of I-PV is considered separate from all of its dependencies. It is lisenced under · Attribution Assurance Lisence.
If you happen to use the software in whole or partially, or reuse subparts of it or get 'inspired' (yes, I hate the word) by it, please do cite the papers below:
REFERENCE(s):
Ibrahim Tanyalcin, Carla Al Assaf, Alexander Gheldof, Katrien Stouffs, Willy Lissens, and Anna C. Jansen. I-PV: a CIRCOS module for interactive protein sequence visualization. Bioinformatics first published online October 10, 2015 doi:10.1093/bioinformatics/btv579
Ibrahim Tanyalcin, Julien Ferte, Taushif Khan and Carla Al Assaf. Indoril: An I-PV Add-On For Visualization Of Point Mutations On 3D Cartesian Coordinates. Biorxiv first posted June 9, 2017 doi:10.1101/148122
I retain the copyrights of Javascript used within this website and I-PV. So if you use parts of the code please give proper credit in a visible section of your project. The output of I-PV is considered separate from all of its dependencies. It is lisenced under · Attribution Assurance Lisence.
If you happen to use the software in whole or partially, or reuse subparts of it or get 'inspired' (yes, I hate the word) by it, please do cite the papers below:
REFERENCE(s):
Ibrahim Tanyalcin, Carla Al Assaf, Alexander Gheldof, Katrien Stouffs, Willy Lissens, and Anna C. Jansen. I-PV: a CIRCOS module for interactive protein sequence visualization. Bioinformatics first published online October 10, 2015 doi:10.1093/bioinformatics/btv579
Ibrahim Tanyalcin, Julien Ferte, Taushif Khan and Carla Al Assaf. Indoril: An I-PV Add-On For Visualization Of Point Mutations On 3D Cartesian Coordinates. Biorxiv first posted June 9, 2017 doi:10.1101/148122
COMMENTS & FEATURE REQUESTS:
This section is intended for feature requests and comments on functionality. (Currently Counts)
This section is intended for feature requests and comments on functionality. (Currently Counts)